Note
Click here to download the full example code
Detect bad sensors using RANSAC¶
This example demonstrates how to use RANSAC 1 from the PREP pipeline to
detect bad sensors and repair them. Note that this implementation in
autoreject 2 is an extension of the original implementation and
works for MEG sensors as well.
References¶
- 1
Bigdely-Shamlo, N., Mullen, T., Kothe, C., Su, K. M., & Robbins, K. A. (2015). The PREP pipeline: standardized preprocessing for large-scale EEG analysis. Frontiers in neuroinformatics, 9, 16.
- 2
Jas, M., Engemann, D. A., Bekhti, Y., Raimondo, F., & Gramfort, A. (2017). Autoreject: Automated artifact rejection for MEG and EEG data. NeuroImage, 159, 417-429.
# Author: Mainak Jas <mainak.jas@telecom-paristech.fr>
# License: BSD (3-clause)
For the purposes of this example, we shall use the MNE sample dataset. Therefore, let us make some MNE related imports.
import mne
from mne import io
from mne import Epochs
from mne.datasets import sample
Let us now read in the raw fif file for MNE sample dataset.
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
raw = io.read_raw_fif(raw_fname, preload=True)
Out:
Opening raw data file /Users/mainak/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Current compensation grade : 0
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
We can then read in the events
event_fname = data_path + ('/MEG/sample/sample_audvis_filt-0-40_raw-'
'eve.fif')
event_id = {'Auditory/Left': 1}
tmin, tmax = -0.2, 0.5
events = mne.read_events(event_fname)
And pick MEG channels for repairing. Currently, autoreject can repair
only one channel type at a time.
raw.info['bads'] = []
Now, we can create epochs. The reject params will be set to None
because we do not want epochs to be dropped when instantiating
mne.Epochs.
raw.info['projs'] = list() # remove proj, don't proj while interpolating
epochs = Epochs(raw, events, event_id, tmin, tmax,
baseline=(None, 0), reject=None,
verbose=False, detrend=0, preload=True)
picks = mne.pick_types(epochs.info, meg='grad', eeg=False,
stim=False, eog=False,
include=[], exclude=[])
We import Ransac and run the familiar fit_transform method.
Out:
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[ ] 0.00% Iterating epochs |
[ ] 1.39% Iterating epochs /
[. ] 2.78% Iterating epochs -
[. ] 4.17% Iterating epochs \
[.. ] 5.56% Iterating epochs |
[.. ] 6.94% Iterating epochs /
[... ] 8.33% Iterating epochs -
[... ] 9.72% Iterating epochs \
[.... ] 11.11% Iterating epochs |
[..... ] 12.50% Iterating epochs /
[..... ] 13.89% Iterating epochs -
[...... ] 15.28% Iterating epochs \
[...... ] 16.67% Iterating epochs |
[....... ] 18.06% Iterating epochs /
[....... ] 19.44% Iterating epochs -
[........ ] 20.83% Iterating epochs \
[........ ] 22.22% Iterating epochs |
[......... ] 23.61% Iterating epochs /
[.......... ] 25.00% Iterating epochs -
[.......... ] 26.39% Iterating epochs \
[........... ] 27.78% Iterating epochs |
[........... ] 29.17% Iterating epochs /
[............ ] 30.56% Iterating epochs -
[............ ] 31.94% Iterating epochs \
[............. ] 33.33% Iterating epochs |
[............. ] 34.72% Iterating epochs /
[.............. ] 36.11% Iterating epochs -
[............... ] 37.50% Iterating epochs \
[............... ] 38.89% Iterating epochs |
[................ ] 40.28% Iterating epochs /
[................ ] 41.67% Iterating epochs -
[................. ] 43.06% Iterating epochs \
[................. ] 44.44% Iterating epochs |
[.................. ] 45.83% Iterating epochs /
[.................. ] 47.22% Iterating epochs -
[................... ] 48.61% Iterating epochs \
[.................... ] 50.00% Iterating epochs |
[.................... ] 51.39% Iterating epochs /
[..................... ] 52.78% Iterating epochs -
[..................... ] 54.17% Iterating epochs \
[...................... ] 55.56% Iterating epochs |
[...................... ] 56.94% Iterating epochs /
[....................... ] 58.33% Iterating epochs -
[....................... ] 59.72% Iterating epochs \
[........................ ] 61.11% Iterating epochs |
[......................... ] 62.50% Iterating epochs /
[......................... ] 63.89% Iterating epochs -
[.......................... ] 65.28% Iterating epochs \
[.......................... ] 66.67% Iterating epochs |
[........................... ] 68.06% Iterating epochs /
[........................... ] 69.44% Iterating epochs -
[............................ ] 70.83% Iterating epochs \
[............................ ] 72.22% Iterating epochs |
[............................. ] 73.61% Iterating epochs /
[.............................. ] 75.00% Iterating epochs -
[.............................. ] 76.39% Iterating epochs \
[............................... ] 77.78% Iterating epochs |
[............................... ] 79.17% Iterating epochs /
[................................ ] 80.56% Iterating epochs -
[................................ ] 81.94% Iterating epochs \
[................................. ] 83.33% Iterating epochs |
[................................. ] 84.72% Iterating epochs /
[.................................. ] 86.11% Iterating epochs -
[................................... ] 87.50% Iterating epochs \
[................................... ] 88.89% Iterating epochs |
[.................................... ] 90.28% Iterating epochs /
[.................................... ] 91.67% Iterating epochs -
[..................................... ] 93.06% Iterating epochs \
[..................................... ] 94.44% Iterating epochs |
[...................................... ] 95.83% Iterating epochs /
[...................................... ] 97.22% Iterating epochs -
[....................................... ] 98.61% Iterating epochs \
[........................................] 100.00% Iterating epochs |[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 2.3min remaining: 0.0s
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 2.3min finished
We can also get the list of bad channels computed by Ransac.
print('\n'.join(ransac.bad_chs_))
Out:
MEG 0722
MEG 0813
MEG 0812
MEG 0822
MEG 0912
MEG 0942
MEG 0943
MEG 1013
MEG 1023
MEG 1022
MEG 1032
MEG 1422
MEG 1633
MEG 2443
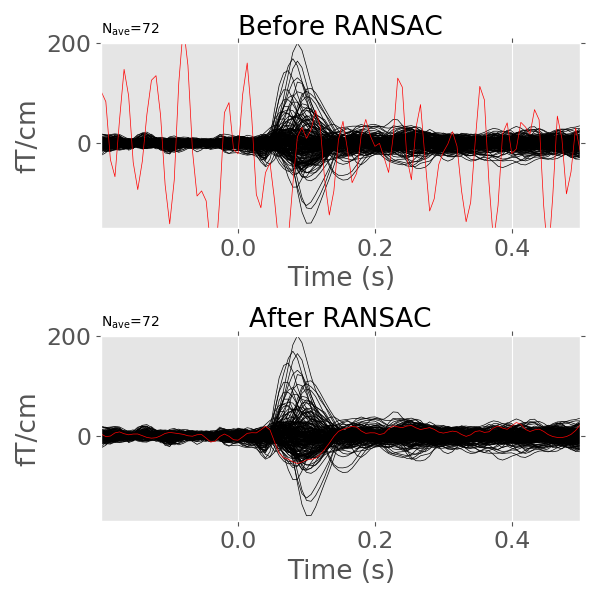
Then we compute the evoked before and after interpolation.
evoked = epochs.average()
evoked_clean = epochs_clean.average()
We will manually mark the bad channels just for plotting.
evoked.info['bads'] = ['MEG 2443']
evoked_clean.info['bads'] = ['MEG 2443']
Let us plot the results.
from autoreject.utils import set_matplotlib_defaults # noqa
import matplotlib.pyplot as plt # noqa
set_matplotlib_defaults(plt)
fig, axes = plt.subplots(2, 1, figsize=(6, 6))
for ax in axes:
ax.tick_params(axis='x', which='both', bottom='off', top='off')
ax.tick_params(axis='y', which='both', left='off', right='off')
ylim = dict(grad=(-170, 200))
evoked.pick_types(meg='grad', exclude=[])
evoked.plot(exclude=[], axes=axes[0], ylim=ylim, show=False)
axes[0].set_title('Before RANSAC')
evoked_clean.pick_types(meg='grad', exclude=[])
evoked_clean.plot(exclude=[], axes=axes[1], ylim=ylim)
axes[1].set_title('After RANSAC')
fig.tight_layout()
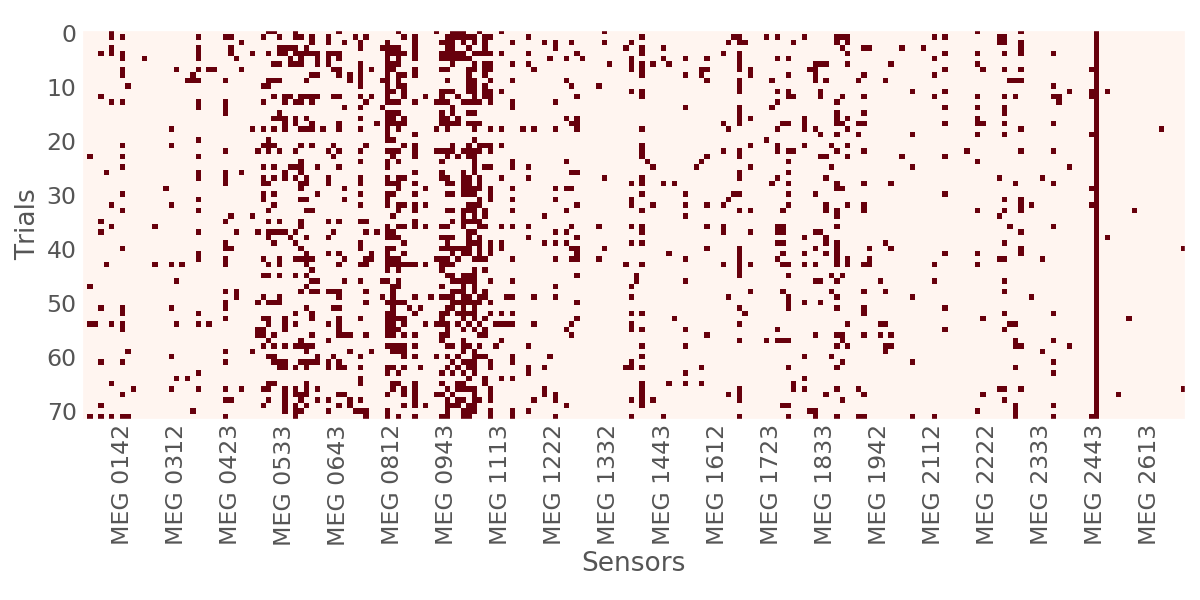
To top things up, we can also visualize the bad sensors for each trial using a heatmap.
ch_names = [epochs.ch_names[ii] for ii in ransac.picks][7::10]
fig, ax = plt.subplots(1, 1, figsize=(12, 6))
ax.imshow(ransac.bad_log, cmap='Reds',
interpolation='nearest')
ax.grid(False)
ax.set_xlabel('Sensors')
ax.set_ylabel('Trials')
plt.setp(ax, xticks=range(7, len(ransac.picks), 10),
xticklabels=ch_names)
plt.setp(ax.get_yticklabels(), rotation=0)
plt.setp(ax.get_xticklabels(), rotation=90)
ax.tick_params(axis=u'both', which=u'both', length=0)
fig.tight_layout(rect=[None, None, None, 1.1])
plt.show()
Out:
/Users/mainak/Documents/github_repos/autoreject/examples/plot_ransac.py:134: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
Total running time of the script: ( 2 minutes 38.312 seconds)
