Note
Go to the end to download the full example code.
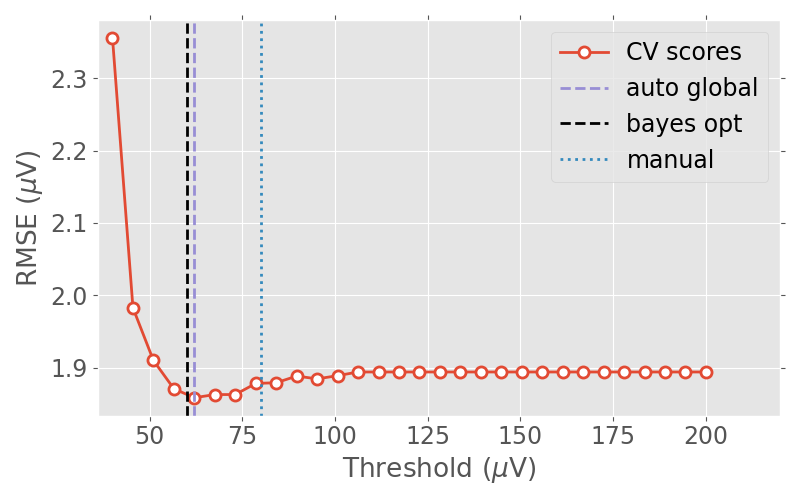
Plotting the cross-validation curve#
This example demonstrates how to use autoreject to
plot the cross-validation curve that is used to estimate the
global rejection thresholds.
# Author: Mainak Jas <mainak.jas@telecom-paristech.fr>
# License: BSD-3-Clause
Let us import the data using MNE-Python and epoch it.
import mne
from mne import io
from mne.datasets import sample
event_id = {'Visual/Left': 3}
tmin, tmax = -0.2, 0.5
data_path = sample.data_path()
meg_path = data_path / 'MEG' / 'sample'
raw_fname = meg_path / 'sample_audvis_filt-0-40_raw.fif'
event_fname = meg_path / 'sample_audvis_filt-0-40_raw-eve.fif'
raw = io.read_raw_fif(raw_fname, preload=True)
events = mne.read_events(event_fname)
include = []
picks = mne.pick_types(raw.info, meg=False, eeg=True, stim=False,
eog=False, include=include, exclude='bads')
epochs = mne.Epochs(raw, events, event_id, tmin, tmax,
picks=picks, baseline=(None, 0),
reject=None, verbose=False, detrend=1)
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
Let us define a range of candidate thresholds which we would like to try. In this particular case, we try from \(40{\mu}V\) to \(200{\mu}V\)
import numpy as np # noqa
param_range = np.linspace(40e-6, 200e-6, 30)
Next, we can use autoreject.validation_curve() to compute the Root Mean
Squared (RMSE) values at the candidate thresholds. Under the hood, this is
using autoreject._GlobalAutoReject to find global (i.e., for all
channels) peak-to-peak thresholds.
from autoreject import validation_curve # noqa
from autoreject import get_rejection_threshold # noqa
_, test_scores, param_range = validation_curve(
epochs, param_range=param_range, cv=5, return_param_range=True, n_jobs=1)
test_scores = -test_scores.mean(axis=1)
best_thresh = param_range[np.argmin(test_scores)]
Using data from preloaded Raw for 73 events and 106 original time points ...
0 bad epochs dropped
We can also get the best threshold more efficiently using Bayesian optimization
reject2 = get_rejection_threshold(epochs, random_state=0, cv=5)
Using data from preloaded Raw for 73 events and 106 original time points ...
Estimating rejection dictionary for eeg
Now let us plot the RMSE values against the candidate thresholds.
import matplotlib.pyplot as plt # noqa
from autoreject import set_matplotlib_defaults # noqa
set_matplotlib_defaults(plt)
human_thresh = 80e-6 # this is a threshold determined visually by a human
unit = r'$\mu$V'
scaling = 1e6
plt.figure(figsize=(8, 5))
plt.tick_params(axis='x', which='both', bottom='off', top='off')
plt.tick_params(axis='y', which='both', left='off', right='off')
colors = ['#E24A33', '#348ABD', '#988ED5', 'k']
plt.plot(scaling * param_range, scaling * test_scores,
'o-', markerfacecolor='w',
color=colors[0], markeredgewidth=2, linewidth=2,
markeredgecolor=colors[0], markersize=8, label='CV scores')
plt.ylabel('RMSE (%s)' % unit)
plt.xlabel('Threshold (%s)' % unit)
plt.xlim((scaling * param_range[0] * 0.9, scaling * param_range[-1] * 1.1))
plt.axvline(scaling * best_thresh, label='auto global', color=colors[2],
linewidth=2, linestyle='--')
plt.axvline(scaling * reject2['eeg'], label='bayes opt', color=colors[3],
linewidth=2, linestyle='--')
plt.axvline(scaling * human_thresh, label='manual', color=colors[1],
linewidth=2, linestyle=':')
plt.legend(loc='upper right')
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 2.308 seconds)